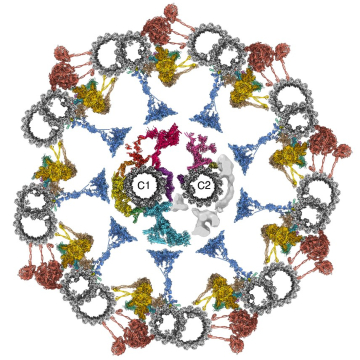
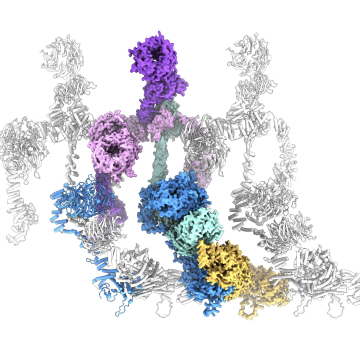
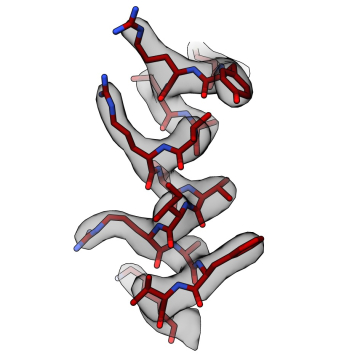
Research
Cilia are conserved and ancient organelles that have been historically underappreciated given their biological importance. These slender, hair-like projections extend from the surface of many eukaryotic cells and have evolved into two main forms: motile and immotile (primary) cilia. Motile cilia are responsible for the locomotion of unicellular eukaryotes and spermatozoa, as well as fluid flow within multicellular organisms. In humans, motile cilia clear mucus from the respiratory tract, circulate cerebrospinal fluid in the brain, and establish asymmetric flow in the embryonic left-right organizer (LRO), setting the left-right body axis. Primary cilia act as antennae for extracellular signals and play important roles in sensory perception and the development and function of many organs and tissues.
Our work aims to determine how cilia are formed, how ciliary signaling pathways are established and maintained, and how motile cilia beat. You can find out more about our research projects by clicking on the links below.
If interested, please consider supporting our research through a gift to our department.